Discover and read the best of Twitter Threads about #3DGenome
Most recents (4)
Ever wonder what the #3Dgenome folding🔺of a Neanderthal looked like?! Could it hold any clues as to where and why we have DNA 🧬and trait differences?
Check out our @capra_lab, Pollard lab & @gfudenberg lab collaboration @biorxivpreprint biorxiv.org/content/10.110…
1/8
Check out our @capra_lab, Pollard lab & @gfudenberg lab collaboration @biorxivpreprint biorxiv.org/content/10.110…
1/8

Sadly, bc of ancient👴sample degradation, we've never explored the role of the 3D genome in archaic hominins like Neanderthals/Denisovans.
But lucky for us, new deep learning models can infer 3D genome structure from DNA sequence alone (like @gfudenberg & @drklly's Akita).
2/8
But lucky for us, new deep learning models can infer 3D genome structure from DNA sequence alone (like @gfudenberg & @drklly's Akita).
2/8

Thrilled to share our latest work, now in @ScienceMagazine! Exploring how #3DGenome organization controls #transcription dynamics during #development. A few highlights below! #InHoxWeTrust
@XinyangBing
Zachary Sisco
@J_Raimundo_
@Michal_Levo
Mike Levine
science.org/doi/10.1126/sc…
@XinyangBing
Zachary Sisco
@J_Raimundo_
@Michal_Levo
Mike Levine
science.org/doi/10.1126/sc…
The Drosophila genome is organized by 2 types of elements: tethers mediate focal contacts, while insulators establish #TAD boundaries. We used genome editing and live transcription imaging to explore their roles in the temporal regulation of #Hox gene expression in the embryo. 

Transcription of the endogenous Scr gene, live! Active transcription foci (white spots) appear in a narrow stripe of nuclei (blue) just prior to the onset of gastrulation. (Scr-MS2, MCP-GFP, His2A-RFP)
Excited to present my first @capra_lab PhD project in today's @GeneticsSociety's @AJHGNews!!
Interested in #3Dgenome structure, complex trait #heritability, and/or evolutionary constraint?
cell.com/ajhg/fulltext/…
1/n
Interested in #3Dgenome structure, complex trait #heritability, and/or evolutionary constraint?
cell.com/ajhg/fulltext/…
1/n
By synthesizing topologically associating domain (TAD) maps across 37 diverse cell types with 41 genome-wide association studies (GWASs), we investigate the differences in disease association and evolutionary pressure on variation across the 3D genome landscape.
2/n
2/n

We know that TAD boundary disruption by SV can lead to developmental disease and cancers. (Thus, selection acts against these SVs).
But what about the relationship between common human variation in TAD boundaries (eg. SNPs) and associations with complex traits?
3/n
But what about the relationship between common human variation in TAD boundaries (eg. SNPs) and associations with complex traits?
3/n
(1/6) Excited to share that our OligoFISSEQ paper is up (nature.com/articles/s4159…)! We set out to develop single cell in situ #3Dgenome mapping methods. A fun collaborative effort led by @huy_sauce @ShyamtanuC and David Castillo, from @TingWu_Lab and @mamartirenom groups. 
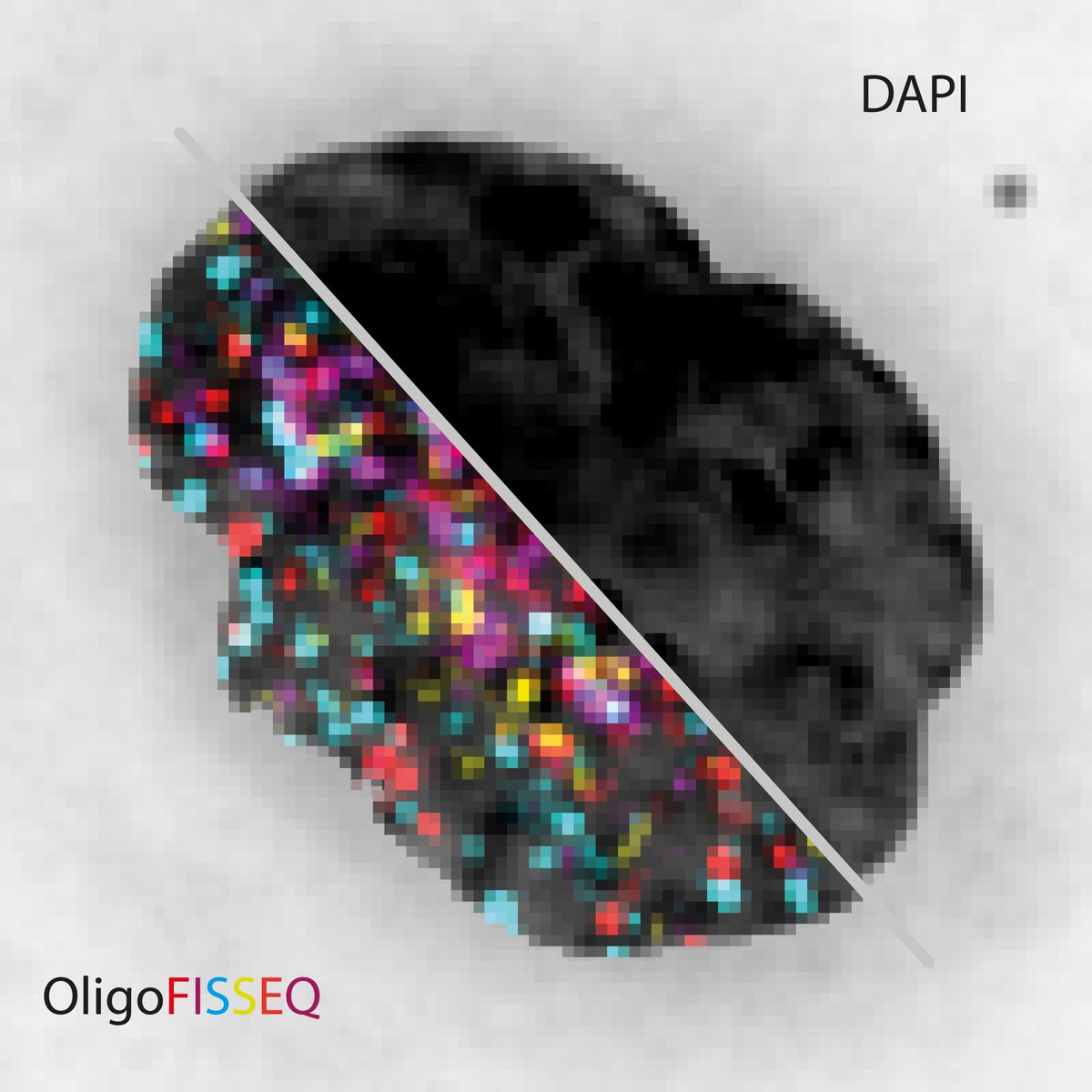
(2/6) An idea that began with @SonCNguyen and Evan Daugharthy. The rest of the team including: @GuyNir5 @AntoniosLioutas @ElliotHershberg @Nuno_MCMartins @PaulReginato Mohammed Hannan @oligopain and @geochurch . Shout out to everyone @naturemethods for the great experience!
(3/6) To target and visualize genomic regions at genome-wide scale, we barcoded Oligopaints, increasing target capacity exponentially. We then visualize and map the targets by #InSituSequencing the barcodes using hybridization, synthesis, and ligation-based approaches.

