Discover and read the best of Twitter Threads about #GWAS
Most recents (20)
Two’s Two’s Two’s - Excited to share our study finding evidence of 2 major subtypes of obesity. Hope these can eventually help support more accurate understanding, diagnosis & treatment of #obesity and #diabetes. go.nature.com/3eNJF1s 
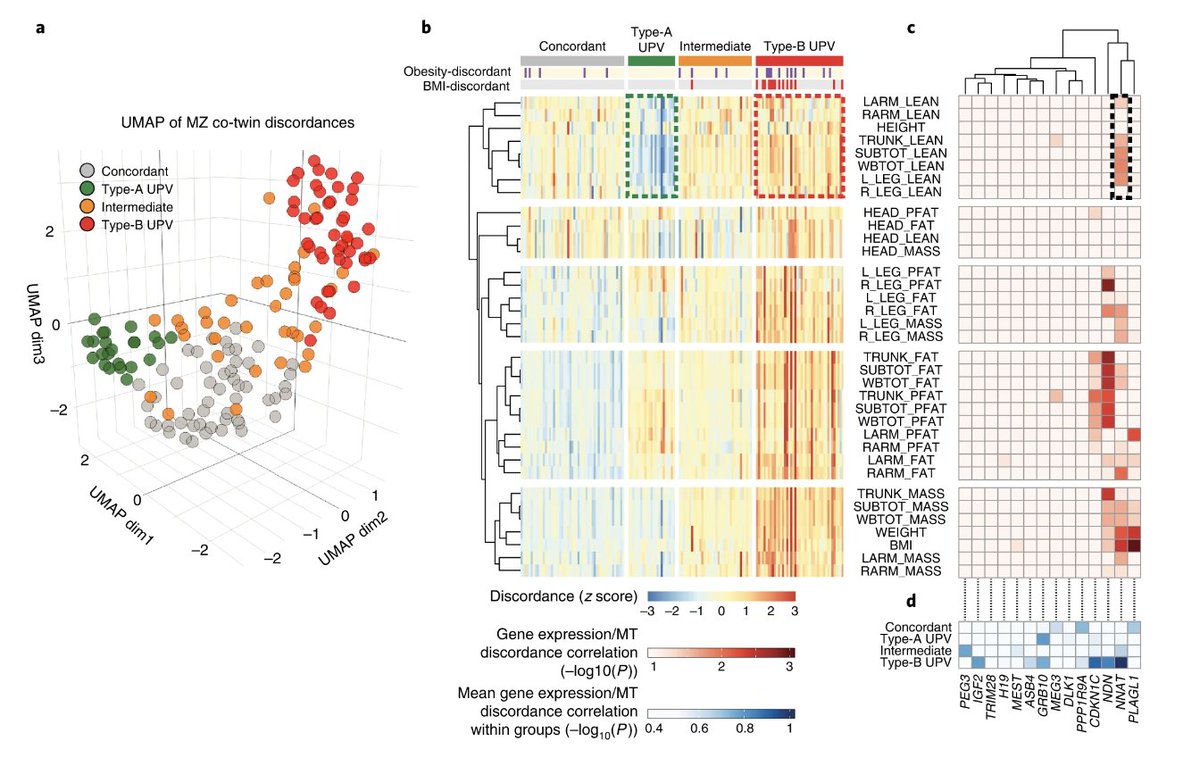
Two patterns of Twin discordance – All started with data-driven finding of 2 major patterns of MZ twin discordance using a dimensional reduction approach. Thank you @TwinsUKres for pioneering cohort and generosity of your staff, funders and participants! nature.com/articles/s4225…
The discordance patterns suggest two forms of human phenotypic plasticity. Type-A = adipose only | Type B = adipose, lean mass, and #insulin co-discordance. Interestingly, Type B shows far more #epigenetics differences suggesting developmental programming.
🚨 Publication alert: largest to date multiancestry exome analysis of body fat distribution discovers INHBE mutations associated with favorable adiposity and PROTECTION from diabetes! 🚨
Out today in @NatureComms
👇🧵 key takeaways below…
nature.com/articles/s4146…
Out today in @NatureComms
👇🧵 key takeaways below…
nature.com/articles/s4146…

Fat distribution: MAJOR BUT OVERLOOKED risk factor for diabetes & heart attacks globally
Storing⬆️fat in waist and⬇️in hips gives ⬆️⬆️⬆️risk – independent of overall body fat (👇)
🚨Lots of drug development in obesity; almost none for fat distribution
nature.com/articles/s4146…
Storing⬆️fat in waist and⬇️in hips gives ⬆️⬆️⬆️risk – independent of overall body fat (👇)
🚨Lots of drug development in obesity; almost none for fat distribution
nature.com/articles/s4146…

Our wolf paper is out! nature.com/articles/s4158… Analysing 72 ancient 🐺 genomes from the last 100,000 years, we: #1 chart wolf natural history through the Ice Age, #2 directly detect natural selection, #3 reveal that dogs have dual ancestry. A 🧵 (11), illustrations by @jessrpeto 

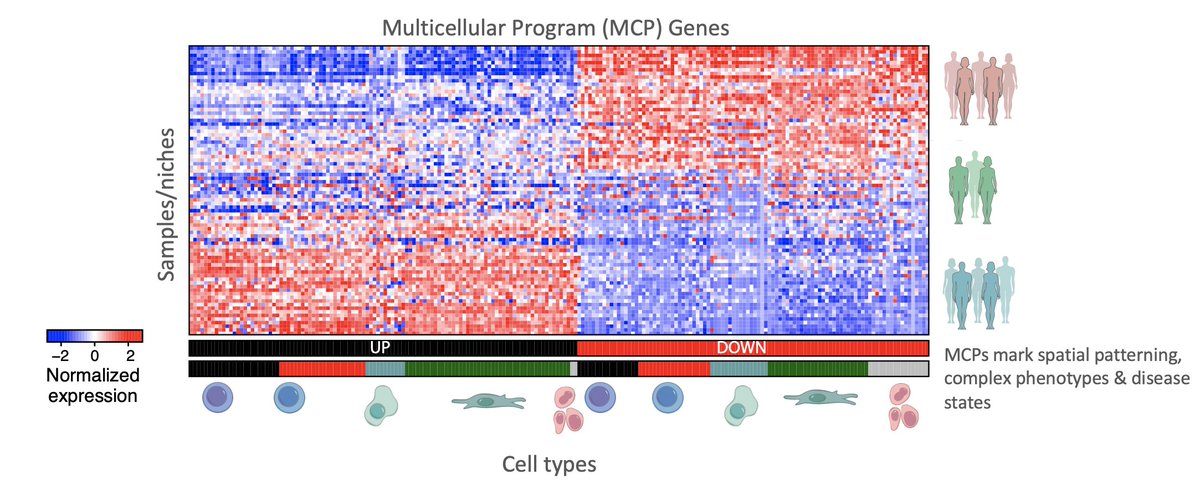
Excited to share that our @NatureBiotech paper with Aviv Regev on Multicellular Programs (MCPs) is now out with a fully automated data-driven method to identify MCPs from #singlecell or spatial data #behindthepaper: go.nature.com/3iQ8fgW nature.com/articles/s4158… (1/19) 

A pinch of pinscher, a dash of 'shund... a pint of pittie for good measure, and what kind of #dog do you get?
Ulti-mutt-ly: a distinct individual.
Happy to share our article on dog genomes, behavior, and MUTTS in @ScienceMagazine! @UMassChan @broadinstitute #ScienceResearch 1/n
Ulti-mutt-ly: a distinct individual.
Happy to share our article on dog genomes, behavior, and MUTTS in @ScienceMagazine! @UMassChan @broadinstitute #ScienceResearch 1/n
Even the most mixy of #mutts cannot escape the question: what's in the sauce? For the first time, we unravel the genomes of mutts (1500+ dogs of mixed or mystery heritage) and what they can show us about the inheritance of behavior. 2/n
Research was made possible thanks to the thousands of dogs enrolled in our Darwin’s Ark project (darwinsark.org). @darwinsarkfnd
See: dog tax attached #communityscience #citizenscience #CitizenScienceMonth 3/n
See: dog tax attached #communityscience #citizenscience #CitizenScienceMonth 3/n
Perceptions of #dog breed temperments are ingrained in our culture. Breed #stereotypes play out in dog parks, show up in city ordinances & leases, are celebrated at dog shows, and infuse Hollywood movies... but what if I told you dog temperment doesn't work like that? 1/n 

ANNOUNCING OUR NEW, AT-LEAST-8-YEARS-IN-THE-MAKING-BUT-WHO'S-COUNTING, +FANCY-GENOMICS #DOG temperment PAPER!!!! @UMassChan @broadinstitute @karlssonlab @darwinsarkfnd #ScienceResearch @iaabc 2/n science.org/doi/10.1126/sc… 

Before I unpack our findings, I have a conflict of interest to disclose: I AM A CAT PERSON. 3/n
Grundlagen der #Pflanzenpathologie. Heute: das #Weizenverzwergungsvirus (WDV). (Auf Anregung von @Fischblog versuchen wir uns mal in einer neuen Twitterkategorie: JKI-Basics) 📜 (1/9) 

Übertragen wird WDV von der #Wandersandzirpe (Psammotettix alienus), einer #Zwergzikade, die in fast ganz Europa verbreitet ist. Sticht sie das #Phloem einer Pflanze an, um Saft zu saugen, dringt das Virus mit dem Speichel in die Pflanze ein. (2/9) 

Die Infektion äußert sich durch eine gestreifte Einfärbung der Blätter, Chlorose (Bleichsucht, häufig Gelbfärbung der Blätter), verringerte Zahl an Ähren, reduzierte Winterfestigkeit und das Absterben von Pflanzen in frühen Entwicklungsstadien. (3/9)
Thrilled to share our work on fine-mapping of immune-mediated diseases (IMD) using regulatory quantitative trait loci (QTLs) finally out in @NatureGenet
nature.com/articles/s4158…
@SoranzoTeam @sangerinstitute @Cambridge_Uni
#immunediseases #autoimmune #GWAS #QTL
🧵 1/14
nature.com/articles/s4158…
@SoranzoTeam @sangerinstitute @Cambridge_Uni
#immunediseases #autoimmune #GWAS #QTL
🧵 1/14
Here we extended the evaluation of regulatory QTLs generated as part of the @blueprint_eu project to systematically map molecular mechanisms and causal variants at 12 immune-mediated diseases with publicly-available summary statistics. 2/14 

📢 The BLUEPRINT "phase 2" genotype data and regulatory QTL summary statistics are available in the @EGAarchive (ega-archive.org/datasets/) under accession codes EGAD00001005192, EGAD00001005199 and EGAD00001005200.
3/14
3/14
Are you interested in how we can learn more human biology by integrating #singlecell genomics and #GWAS?
Please check out our preprint:
#V2F mapping at single-cell resolution through network propagation
biorxiv.org/content/10.110…
Led by @fulong_yu! A short 🧵 (1/n)
Please check out our preprint:
#V2F mapping at single-cell resolution through network propagation
biorxiv.org/content/10.110…
Led by @fulong_yu! A short 🧵 (1/n)

#GWAS have identified innumerable variants associated with disease, as shown in this recent @GWASCatalog plot, yet our understanding of the function of most variants is lacking. #V2F (2/n) 

With the increasing availability of #singlecell genomic atlases, such as the @humancellatlas, there are tremendous opportunities to systematically map variants to functional regulatory elements, which can be defined by accessible chromatin or epigenomic marks. (3/n)
A great paper by Dr. @SaraSuliman13 on her work with Dr. Luo on #tuberculosis in #Peruvians: nature.com/articles/s4146… #BIIW21 #BIIAtTheBench
#GWAS can be hard to link genes together and that's okay. This means we need to regroup #BIIW21 #BIIAtTheBench
Really impressive talk on noncoding #constraint in #gnomAD genomes from Siwei Chen (@konradjk's lab).
#ASHG21
#ASHG21
Chen: Calculated constraint on 1kb windows using Z scores.
How do known non-coding elements (like enhancers) look when viewed through the lens on constraint?
#ASHG21
How do known non-coding elements (like enhancers) look when viewed through the lens on constraint?
#ASHG21
Chen: When looking at largest constraint Z-scores (top Z-score was 4 on the figures)
- Super enhancers ~3x enriched
- ENCODE cCRE enhancers ~2.25x enriched
- FANTOM enhancers ~1.75x enriched
#ASHG21
- Super enhancers ~3x enriched
- ENCODE cCRE enhancers ~2.25x enriched
- FANTOM enhancers ~1.75x enriched
#ASHG21
Excited to share our new #method, scDRS, for associating **individual cells** to disease by integrating #scRNA-seq + #GWAS, providing #polygenic disease risk enrichment beyond tissues or cell types.
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
scDRS assesses excess expression of a set of putative disease genes constructed from GWAS, using an appropriately matched empirical null distribution to compute p-values. (2/n) 

We applied scDRS to GWAS from 74 diseases and complex traits (average N=341K) in conjunction with 16 scRNA-seq data sets spanning 1.3 million cells from 31 tissues and organs, including the amazing Tabula Muris Senis mouse atlas and Tabula Sapiens human atlas @czbiohub. (3/n) 

Extremely proud to share our study on pleiotropy and multi-trait #GWAS recently published in @PLOSGenetics
journals.plos.org/plosgenetics/a…
Here is a brief summary of the findings:
journals.plos.org/plosgenetics/a…
Here is a brief summary of the findings:
We compared several multi-trait tests on real and simulated datasets. In most situations, the test referred as Omnibus in our study turn out to have more statistical power.
Using multi-trait tests on publicly available GWAS summary statistics (36 phenotypes), we uncovered 322 new associations.
1/n Super proud of work published this week by @anzheng25 et al. in @NatMachIntell using #deeplearning to identify sequence context features predictive of transcription factor binding. rdcu.be/cdMmE Some key points:
2/n The main idea: TFs typically bind short motifs of 6-12bp. But only a small fraction of motifs in the genome are actually bound. How well can the question of “to bind or not to bind” be predicted by sequence context (1kb) around the motif using #DeepSea style CNNs?
3/n Pretty well! For most TFs we tried, we could predict whether its motif was bound based on ChIP-seq very well (mean auROC ~0.94) just from local sequence context
How do the hundreds of genetic risk-factors for #Schizophrenia actually lead to disease? New work medrxiv.org/content/10.110… from @manoliskellis illuminates the #SingleCell landscape of #Brains from patients with #Schizophrenia and their healthy counterparts🔬🧠🧵1/10
Combining single-nuclear RNA-Seq @10xGenomics, barcode #ing, and multi-level cell-state decomposition, first author Brad Ruzicka et al. identify 20 cell types in the prefrontal cortex of 24 patients with #Schizophrenia and 24 controls 2/10 
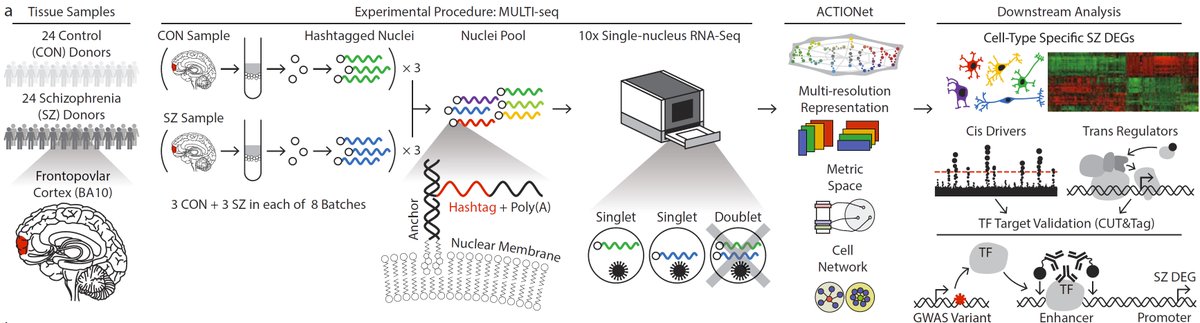
While most of these #cells have been described before, what's powerful about the approach is that we can now ask: are there specific cell-types that are increased or decreased in numbers in #Schizophrenia patients compared to healthy individuals (great study design) 3/10 

Immensely proud of bring of bringing some important aspects of #precision #proteomics to @NatureRevGenet with @ksuhre @markmccarthyoxf covering #gwas #pqtls and more nature.com/articles/s4157… @scilifelab @KTHresearch @ProteinAtlas
We provide insights into procedures, challenges, considerations and opportunities when combing #genetics and #proteomics for more informed, large scale studies of the circulating #proteome . #biobanks #olink #somalogic #massspec #affinityproteomics #epitopes #causality
Am very grateful to @ksuhre and @markmccarthyoxf for joining in on this journey and making this a wonderful experience over three continents, timezones, project and travel schedules!
I found the discussion of Quetelet and populations at the beginning of Chapter 3 of Epidemiology and the People’s Health particularly instructive, especially for genetic epidemiology. #epibookclub #epipeopleshealth #gwas #genepitwitter 1/18 


The question “who—or what—determines populations or groups that merit comparison” is an important but tetchy one. 2/18
The concept of “population stratification bias” in genetic epidemiology is usually introduced using a toy example: say we’re studying two populations, with random mating within but no mating across populations. 3/18
@my_helix @uk_biobank @my_helix took a look at 1,317 @uk_biobank phenotypes w/ 50,000 exomes released last week. Going beyond #GWAS, used gene-based collapsing analysis. Thanks @bmneale Pheasant pipeline for clean phenotypes, @hailgenetics for the tooling.
@my_helix @uk_biobank @bmneale @hailgenetics Found some stuff that we expected, #LDLR for #cholesterol, #MC1R for red hair; but counts too small for other known conditions like HBOC/#BRCA2 @ATorkmani @RobertCGreen @atulbutte @gabecasis
@my_helix @uk_biobank @bmneale @hailgenetics @atorkmani @RobertCGreen @atulbutte @gabecasis Some interesting findings: #GP1BB associated with mean platelet volume. #GP1BB variants have been implicated in bleeding and platelet disorders. 
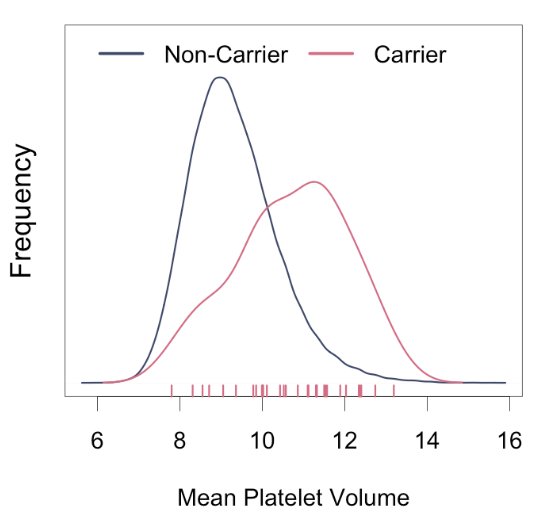
@f2harrell @NPirastu @paulpharoah I worry three very distinct scientific goals for #GWAS and genetic association studies broadly are being conflated on this thread: (1) locus discovery, (2) causal variant discovery, and (3) disease prediction. Each requires different analysis strategies and interpretation. 1/n
@f2harrell @NPirastu @paulpharoah As to (1), as @tamar_sofer @paulpharoah and others have noted, the goal is just to find markers that are correlated with a causal variant (or variants). Nobody claims—or nobody should be claiming—that these markers are unique. 2/n
@f2harrell @NPirastu @paulpharoah @tamar_sofer So it’s not surprising if Study A reports SNP 1 at Locus X and Study B reports SNP 2. Nor is it particularly disturbing, if SNPs 1 & 2 are correlated. If you dig thru supplements you will probably find that SNP 2 has strong evidence for association in Study A and vice versa. 3/n
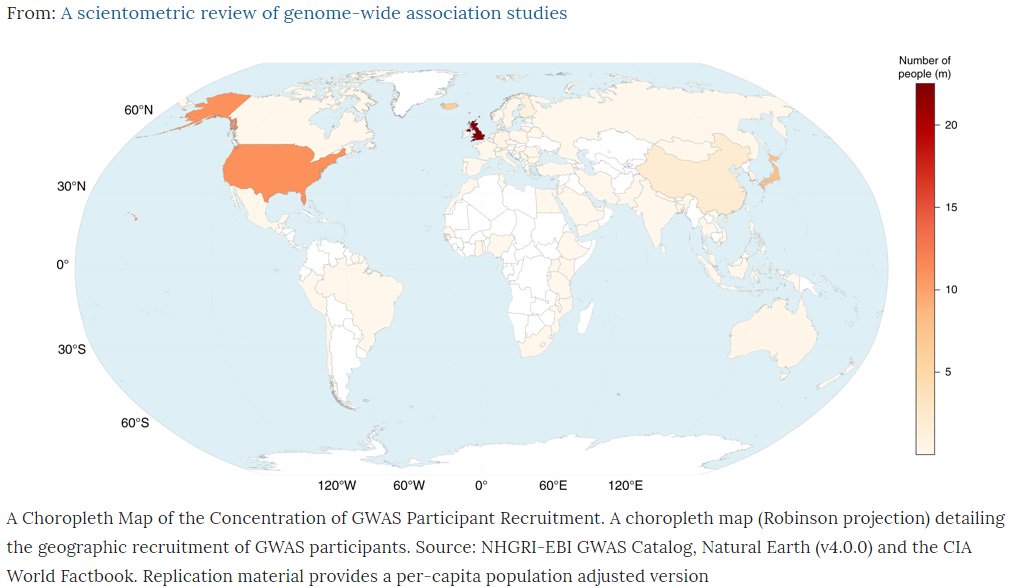
Published open access today @CommsBio our review of all ~4000 #GWAS (genetic discoveries) from @GWASCatalog nature.com/articles/s4200… 1/9 
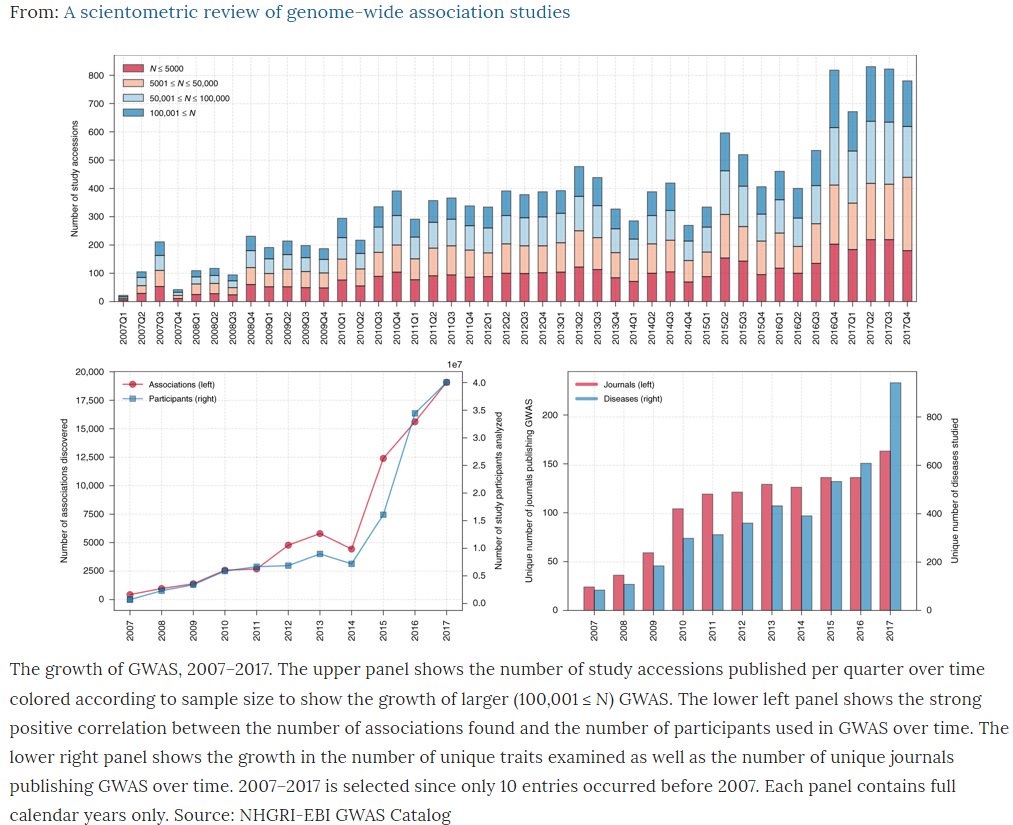
Although explosion in number of #GWAS studies, sample sizes, associations and diseases – genetic discovery still largely of European ancestry subjects – when non-European groups included more often for replication 2/9 
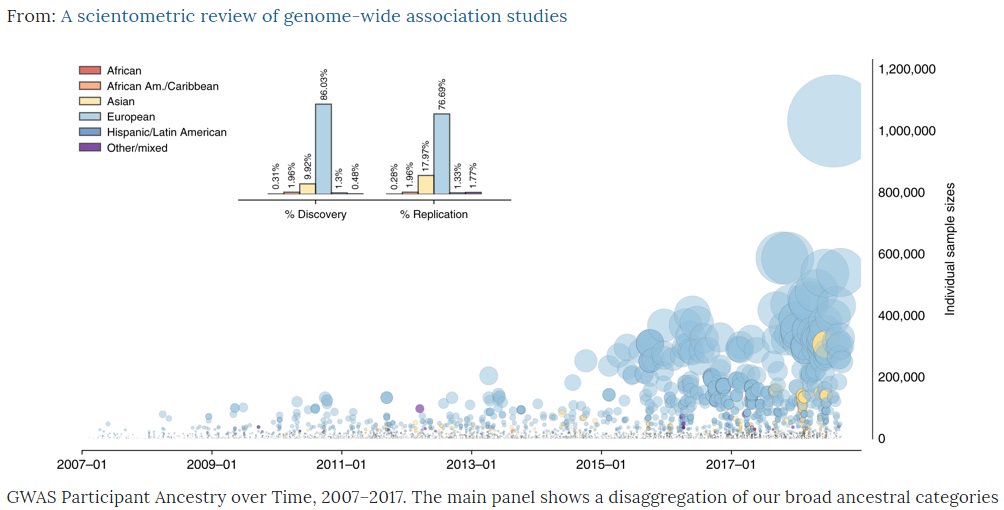
A world map of genetic discoveries shows 72% of subjects in #GWAS studies come from just 3 countries – US, UK and Iceland 3/9