Discover and read the best of Twitter Threads about #SundayMultiOmics
Most recents (3)
PathME unsupervised #multiomics
1) genes space → pathways space
2) for each pathway: collapse pathways from multiple omics to one per patient
3) sparse NMF biclustering
✓compared against SNF and iCluster
✓TCGA x 4
✓hyperparameters
✓source code
✓5-fold CV
#SundayMultiOmics
1) genes space → pathways space
2) for each pathway: collapse pathways from multiple omics to one per patient
3) sparse NMF biclustering
✓compared against SNF and iCluster
✓TCGA x 4
✓hyperparameters
✓source code
✓5-fold CV
#SundayMultiOmics
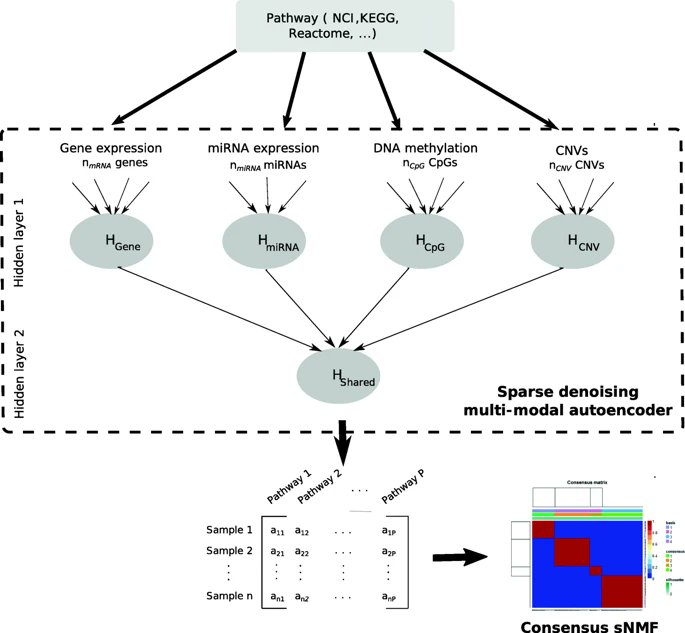
Article: doi.org/10.1186/s12859…
GitHub: github.com/AminaLEM/PathME
Figure © by authors, reused under CC-BY 4.0:
creativecommons.org/licenses/by/4.0
#omics #NeuralNetworks
GitHub: github.com/AminaLEM/PathME
Figure © by authors, reused under CC-BY 4.0:
creativecommons.org/licenses/by/4.0
#omics #NeuralNetworks
Worth noting:
- authors use sNMF consensus from 500 runs (cophenetic correlation + permutation testing to choose # of clusters)
- the autoencoders are denoising
- worth praise is the effort into interpretability (of both features/omics & clinical associations) - see supplement!
- authors use sNMF consensus from 500 runs (cophenetic correlation + permutation testing to choose # of clusters)
- the autoencoders are denoising
- worth praise is the effort into interpretability (of both features/omics & clinical associations) - see supplement!
Multi-Omic inTegrative Analysis (MOTA): an application of differential network analysis to #multiomics.
✓3 non-TCGA datasets (HCC vs CIRR)
✓explanations for parameters & rgCCA choice
✓good at known cancer drivers recovery
✓consistent across cohorts
#SundayMultiOmics 1/n


✓3 non-TCGA datasets (HCC vs CIRR)
✓explanations for parameters & rgCCA choice
✓good at known cancer drivers recovery
✓consistent across cohorts
#SundayMultiOmics 1/n
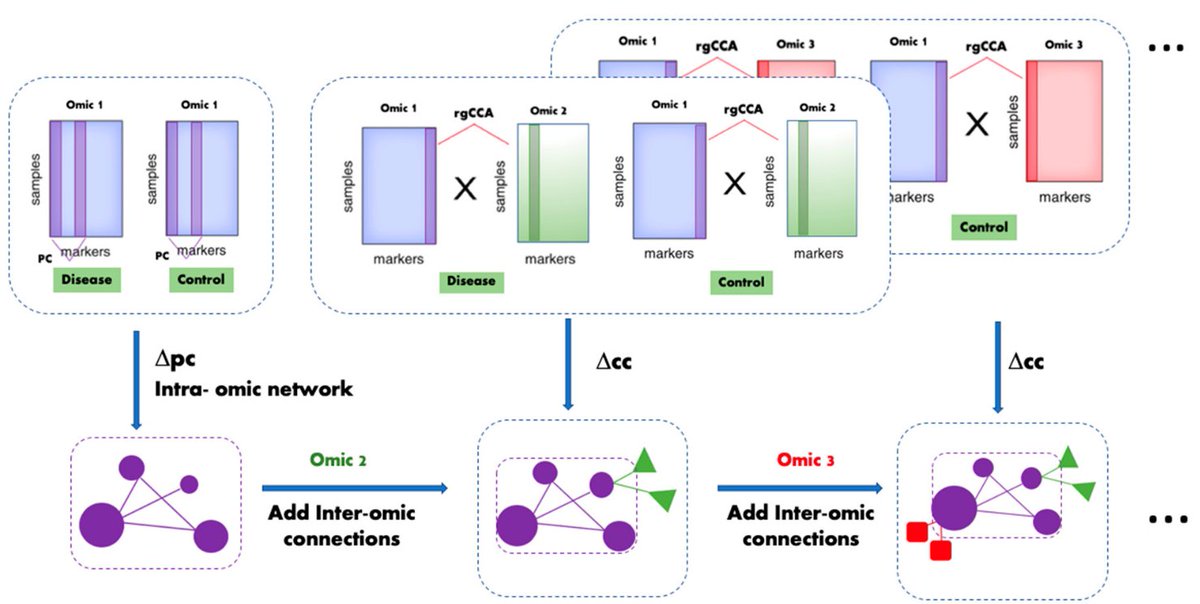
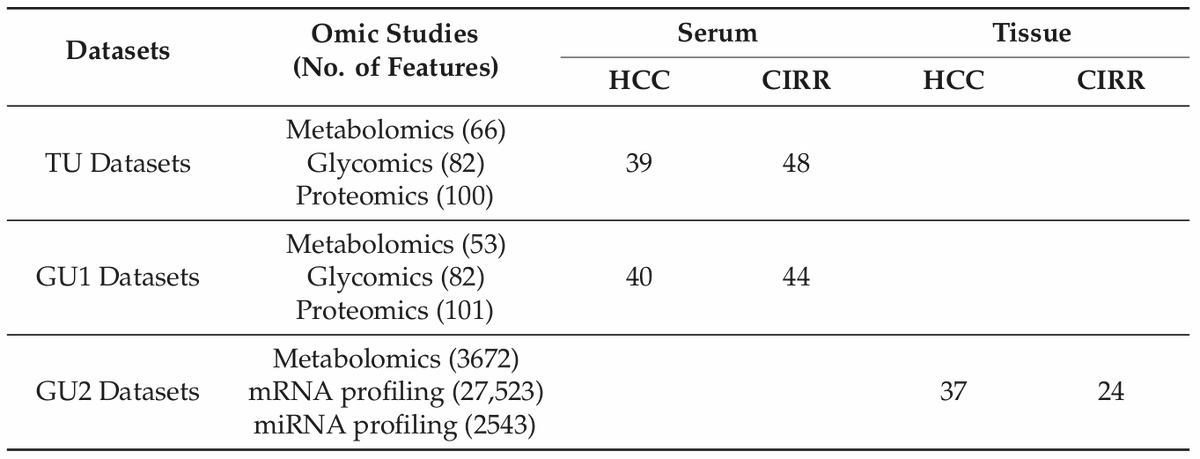
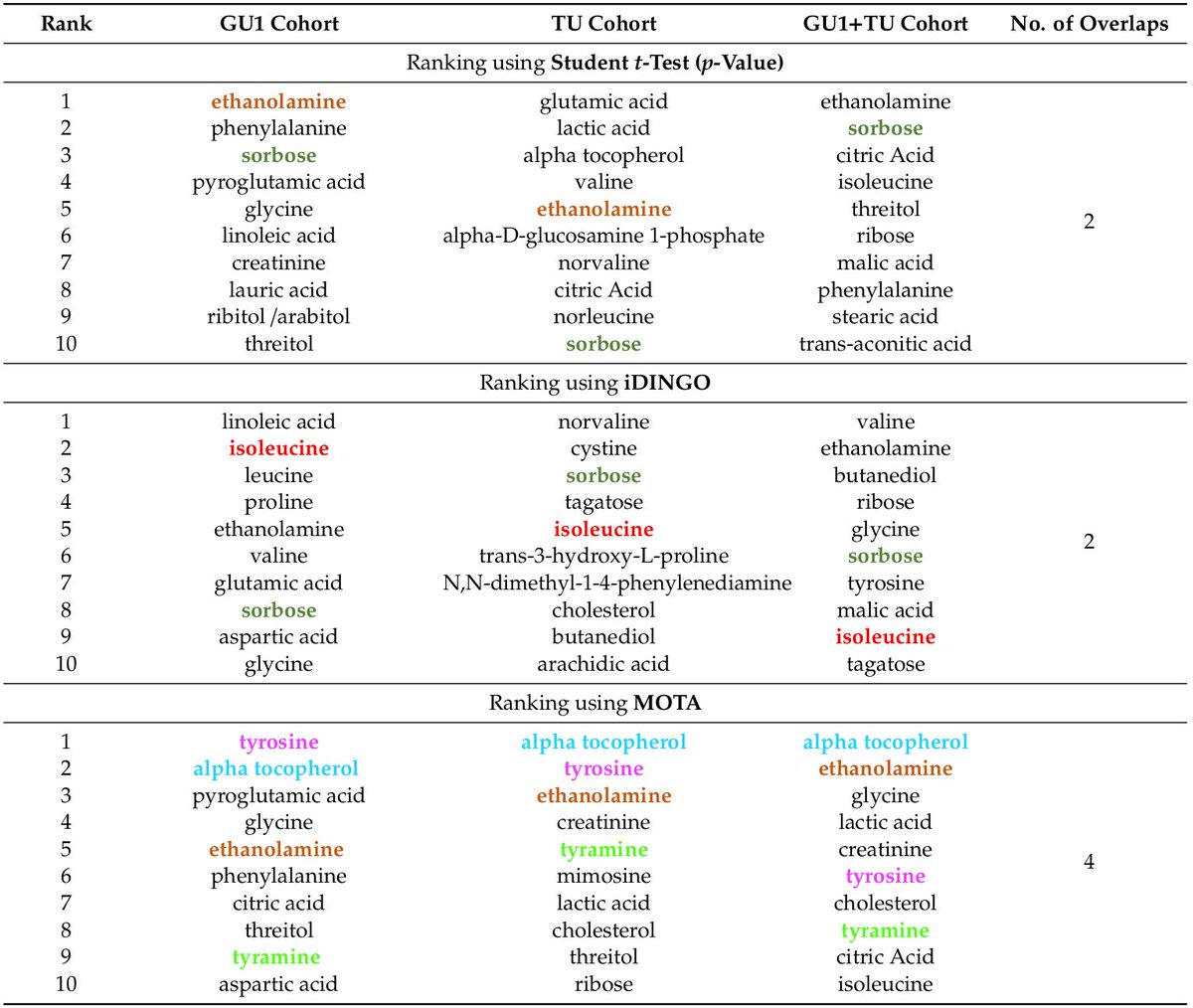
[[ Detailed description & comments follow ]]
Link: doi.org/10.3390/metabo…
Figures © by authors, reused under CC-BY 4.0
creativecommons.org/licenses/by/4.…
Developed @LombardiCancer @gumedcenter (sorry I could not to find authors on Twitter)
Link: doi.org/10.3390/metabo…
Figures © by authors, reused under CC-BY 4.0
creativecommons.org/licenses/by/4.…
Developed @LombardiCancer @gumedcenter (sorry I could not to find authors on Twitter)
["Layman" introduction]: many studies focus on the differences in the abundance of specific biomolecules; while very useful to highlight the obvious #biomarkers, it may not explain complex disease mechanisms & can be very sensitive to cohort differences (e.g. in #metabolomics)
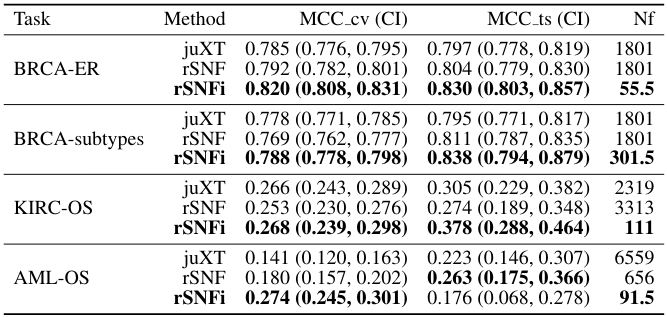
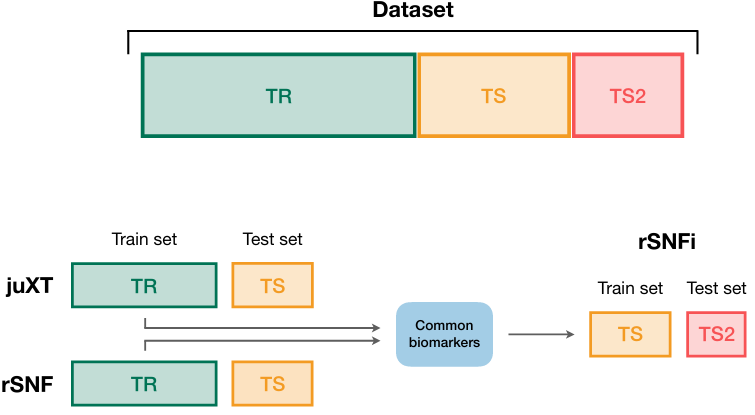
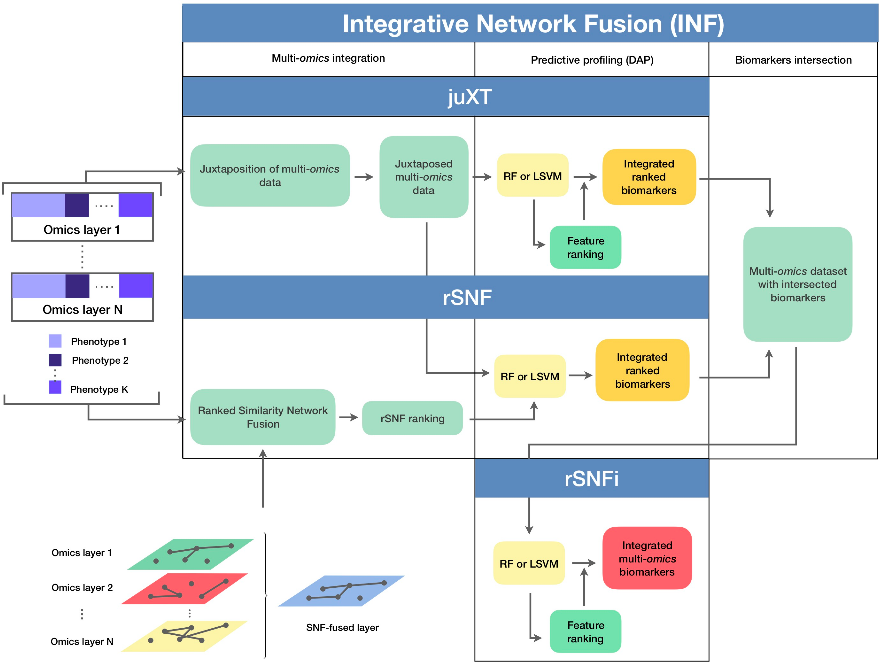
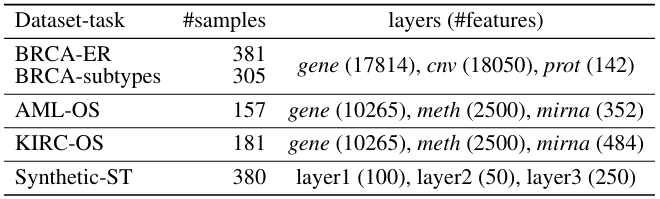
A true gem among #multiomics preprints: Integrative Network Fusion by @MarcoChierici, @nicole_bussola, @viperale, et al:
✓ 3 TCGA cancers & simulated data
✓ cross-validation described in detail
✓ flow diagram
✓ source code & data shared
✓ packages w/ version, cited
/n



✓ 3 TCGA cancers & simulated data
✓ cross-validation described in detail
✓ flow diagram
✓ source code & data shared
✓ packages w/ version, cited
/n
- [the method description & comments follows]
- link: biorxiv.org/content/10.110…
- licence the above figures/tables: CC BY-NC-ND 4.0
- an earlier version of INF was previously presented in 2018: doi.org/10.1186/s13062…
- this is the first tweet in #SundayMultiOmics series
- link: biorxiv.org/content/10.110…
- licence the above figures/tables: CC BY-NC-ND 4.0
- an earlier version of INF was previously presented in 2018: doi.org/10.1186/s13062…
- this is the first tweet in #SundayMultiOmics series
[[Introduction]]: Similarity network fusion (SNF, doi.org/10.1038/nmeth.…) is a popular technique (600+ citations, a lot for multi-omics!) for getting a sort of consensus signal from multiple omics; it requires the same patients (less commonly - observations) in each omic.
