Discover and read the best of Twitter Threads about #DESeq2
Most recents (3)
1/ Are you a bioinformatics researcher looking for powerful tools to analyse your data? Check out @Bioconductor ! Here are some of my favorite packages for #bioinformatics analyses. 

2/ First up: #DESeq2 by @mikelove. This package provides a method for differential gene expression analysis of RNA-seq data. It's widely used and highly cited in the field, and it's perfect for identifying genes that are differentially expressed between samples.
3/ Next, I recommend #edgeR. Like DESeq2, edgeR is a package for differential gene expression analysis of RNA-seq data. It's particularly useful for smaller sample sizes and can detect differential expression with greater sensitivity.
This #EOSS funding from @cziscience for #DESeq2 and #tximeta wrapped up at the end of 2021.
Reporting in this 🧵 on what we developed:
Reporting in this 🧵 on what we developed:
1. @kwame_forbes wrote DESeq2::integrateWithSingleCell() which helps user locate publicly available SC datasets followed by visualization with his own R package:
kwameforbes.github.io/vizWithSCE/
Kwame was then a @UNCPREP scholar, now a first year BCB student at UNC 🧬💻🎉
kwameforbes.github.io/vizWithSCE/
Kwame was then a @UNCPREP scholar, now a first year BCB student at UNC 🧬💻🎉

2. Some Bioc folks and a team at UNC worked on extending the tximeta + DESeq2 + plyranges workflow that @_StuartLee @lawremi and I started in the fluentGenomics paper:
sa-lee.github.io/fluentGenomics/
sa-lee.github.io/fluentGenomics/

Multi-Omic inTegrative Analysis (MOTA): an application of differential network analysis to #multiomics.
✓3 non-TCGA datasets (HCC vs CIRR)
✓explanations for parameters & rgCCA choice
✓good at known cancer drivers recovery
✓consistent across cohorts
#SundayMultiOmics 1/n


✓3 non-TCGA datasets (HCC vs CIRR)
✓explanations for parameters & rgCCA choice
✓good at known cancer drivers recovery
✓consistent across cohorts
#SundayMultiOmics 1/n
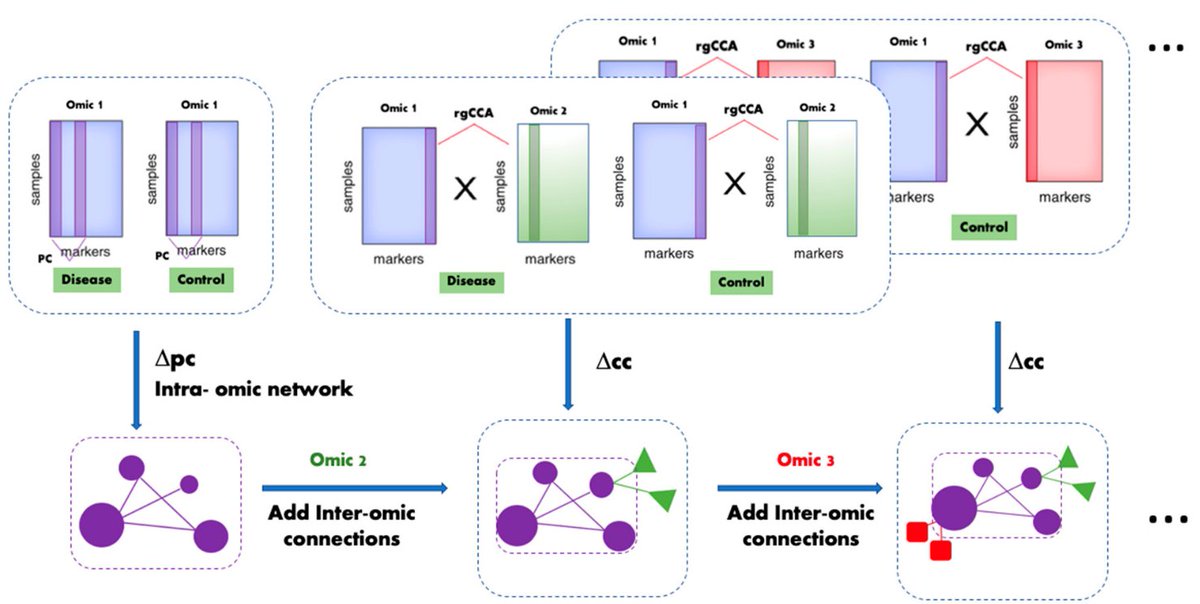
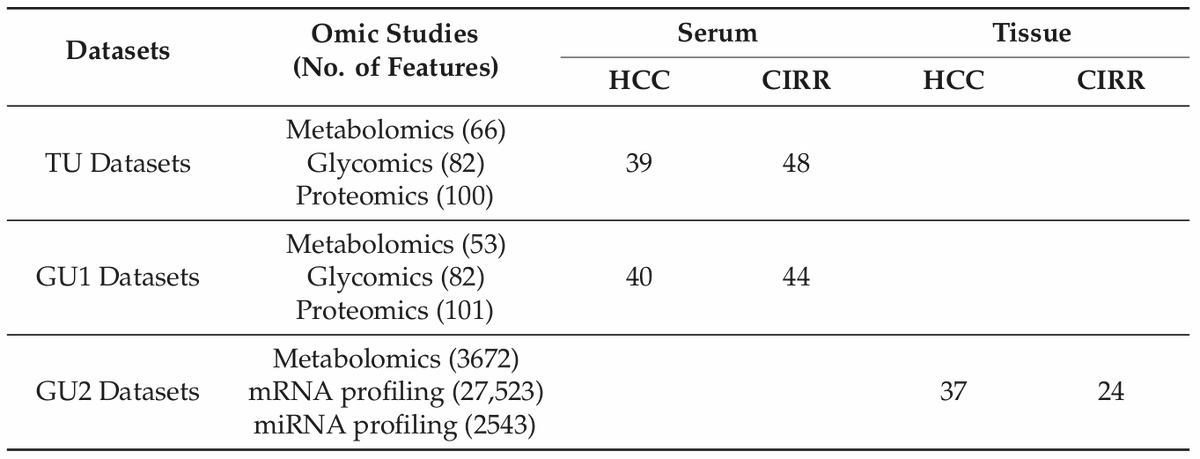
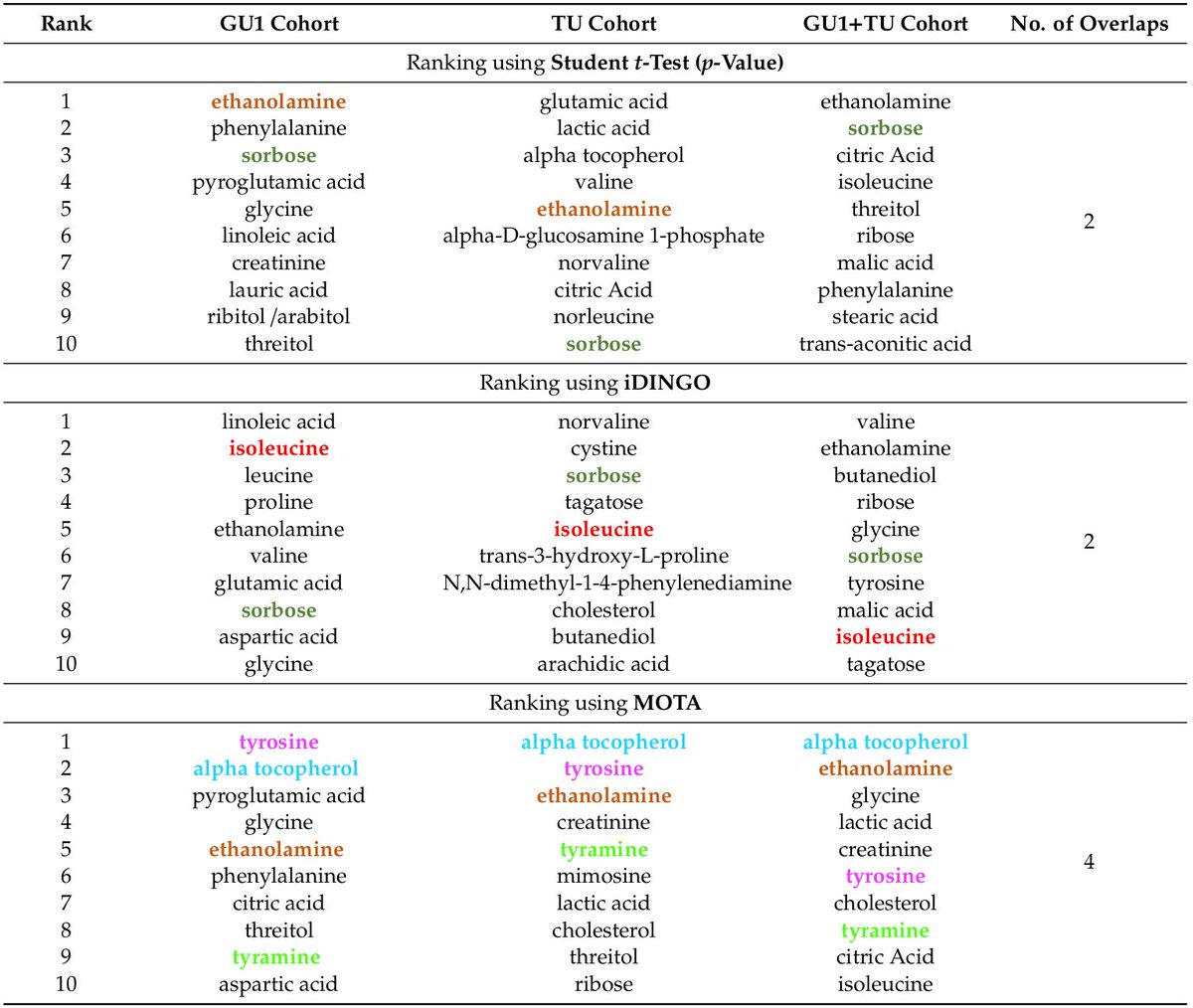
[[ Detailed description & comments follow ]]
Link: doi.org/10.3390/metabo…
Figures © by authors, reused under CC-BY 4.0
creativecommons.org/licenses/by/4.…
Developed @LombardiCancer @gumedcenter (sorry I could not to find authors on Twitter)
Link: doi.org/10.3390/metabo…
Figures © by authors, reused under CC-BY 4.0
creativecommons.org/licenses/by/4.…
Developed @LombardiCancer @gumedcenter (sorry I could not to find authors on Twitter)
["Layman" introduction]: many studies focus on the differences in the abundance of specific biomolecules; while very useful to highlight the obvious #biomarkers, it may not explain complex disease mechanisms & can be very sensitive to cohort differences (e.g. in #metabolomics)
