Discover and read the best of Twitter Threads about #methylation
Most recents (11)
1/5 Fascinating work from Jae-Hyun Yang and colleagues on #mammalian #aging, published in Cell...
An abstract thread 🧵...
"All living things experience an increase in #entropy, manifested as a loss of #genetic and #epigenetic information."
cell.com/cell/fulltext/…
An abstract thread 🧵...
"All living things experience an increase in #entropy, manifested as a loss of #genetic and #epigenetic information."
cell.com/cell/fulltext/…
2/5 "In #yeast, #epigenetic information is lost over time due to the relocalization of #chromatin-modifying proteins to #DNA breaks, causing cells to lose their identity, a hallmark of yeast #aging."
3/5 "Using a system called “#ICE” (#inducible changes to the #epigenome), we find that the act of faithful #DNA #repair advances aging at #physiological, #cognitive, and #molecular #levels, including #erosion of the #epigenetic #landscape,..."
As 2022 draws to a close, we’ve been reflecting on some of the highlights (in no particular order) — you can read the full article here: bit.ly/3IfaKYc, or digest the thread below. Wishing you a happy new year and all the best for 2023! 1/23
The #cancer research community has made huge strides leveraging the unique attributes of #nanopore sequencing to characterise cancer at unprecedented resolution, unlocking previously hidden variation and accelerating research to support human health. nanoporetech.com/cancer-research 2/23
In May, @mdelledon took a group of scientists into the Gobi Desert to capture small mammals, including bats, to study their microbiome and they set up a genomics lab in a tent. They demonstrated the utility of the MinION in the harsh, off grid and sandy desert environment! 3/23
Have you ever wished to sequence SPECIFIC bacteria but NOT the vast background in #microbiome? Today, we introduce *mEnrich-seq*: #methylation-guided enrichment sequencing of bacterial taxa of interest from microbiome. 4yr project led by @clannabel7! biorxiv.org/content/10.110…🧵1/10 

#Metagenomic sequencing throughput is largely consumed by abundant microbes while those with relatively lower abundance cannot be sequenced in a cost-effective manner, due to the highly skewed abundance distribution of diverse bacteria and host DNA. 2/10
Although several methods have been developed (pls see Introduction in preprint) to exclude host DNA and/or the highly abundant microbes, there are still key challenges in achieving high enrichment efficiency and differentiating between bacterial taxa w/ very similar genomes. 3/10
Time for the latest @nanopore technology updates. Up next in the auditorium: an Update from the Oxford Nanopore team. #nanoporeconf 

Please note, we invite you to read the disclaimer in the slides
CB: More information on Clive’s previous technology updates:
10 minute reminder: @VanessaPorter will be on stage in the auditorium shortly with her talk: 'Identification of novel #genomic structures and regulation patterns at #HPV integration events in cervical #cancer'. #nanoporeconf 

VP: HPV is a necessary driver of cervical cancer. HPV integration is present 70% of cervical cancers. Often disrupts E2 gene & causes structural rearrangements. #nanoporeconf
VP: HPV integration often upregulated neighbouring genes. #nanoporeconf
Epigenomic biomarkers are becoming more established for #LiquidBiopsy and #CancerScreening, and we have seen the big players positioning themselves in this #epigenomics race recently.
Catching signs of #cancer early is crucially important to the disease management and survival rates, so the question is: how can we find out if there is something wrong going on early enough, ideally in a low-cost assay that can be performed regularly on healthy individuals?
@sbarnettARK Epigenomic biomarkers are becoming more established for #LiquidBiopsy and #CancerScreening, and we have seen the big players positioning themselves in this #epigenomics race recently.
@sbarnettARK Catching signs of #cancer early is crucially important to the disease management and survival rates, so the question is: how can we find out if there is something wrong going on early enough?
@sbarnettARK The first generation of high-throughput technologies was predicated on finding mutated (tumor) DNA in the individual's body (somatic), different from their (normal) DNA. Tumor/Normal (T/N) comparisons of the individual's samples, biopsies or ctDNA will indicate if there
Interesting that Exact acquired Ashion (private) ostensibly to bring tumor-normal profiling onto the platform. This comes after they got exclusive rights to the TARDIS platform for MRD testing. Both Ashion and TARDIS come out of @TGen, so likely a smoother integration.
Here's the original TARDIS paper: tgen.org/news/2019/augu…
Seems to be another form of error-corrected sequencing, which is useful for liquid biopsy applications where tumor fraction (amount of cancer in body) is very low, so this reads through to mutation-based early ...
Seems to be another form of error-corrected sequencing, which is useful for liquid biopsy applications where tumor fraction (amount of cancer in body) is very low, so this reads through to mutation-based early ...
... detection assays (CancerSEEK) as well as for MRD, where tumor burden is low, but more emphasis on dynamics of the tumor (treatment resistance, clonal evolution, etc) in addition to just quantification of the tumor (ctDNA % up or down).
#JPM2021 @TwistBioscience My Highlights: back to the #DNAWrite field. TwistBio chip: 1M oligos but only as centralised factory setting. 

Expanding to a new factory in Portland (roadmap 2022) to reduce TAT. Also mentioned the "long tail" of Clonal Ready Gene Fragments. 

They have a few slides on #DNAasStorage with a denser chip in prototype phase where they think they can archive 1Tb for $100 (pay once, archive forever*) which, if my calculations are correct, would allow for 10-12 years of @awscloud Deep Glacier or similar mth/GB=0.00099*12*1E3 

#JPM2021 $GH #GuardantHealth my highlights: jpmorgan.metameetings.net/events/healthc… #LiquidBiopsy they are at the forefront of #Epigenomic profiling (Methylation+Fragmentomics) 

#LUNAR1 #CRC bringing #Epigenomic profiling to recurrence testing with #Methylation profiling (no fragmentomics?) 

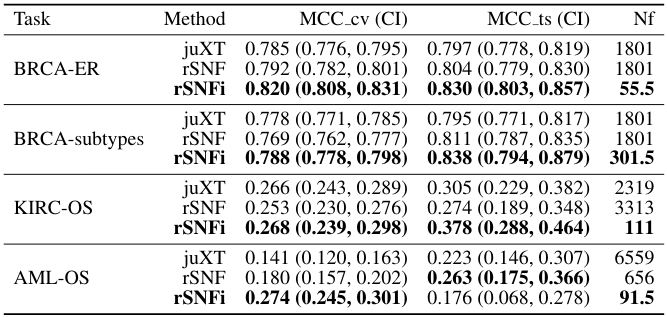
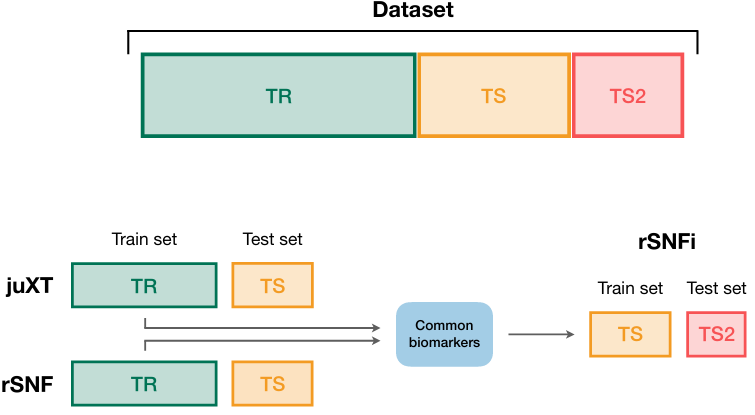
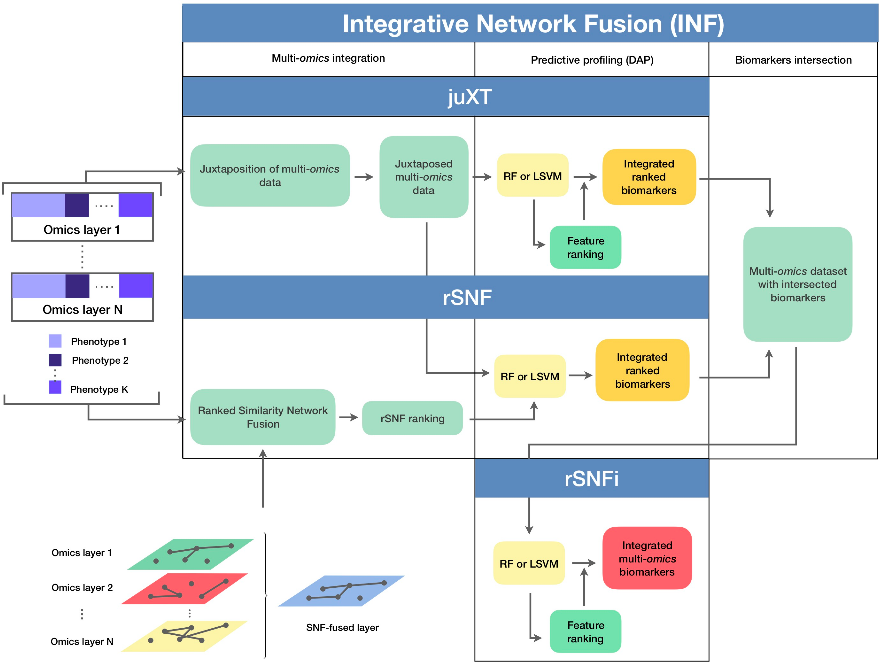
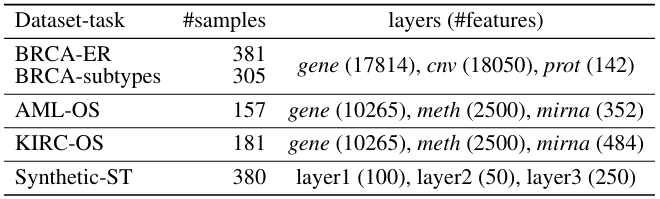
A true gem among #multiomics preprints: Integrative Network Fusion by @MarcoChierici, @nicole_bussola, @viperale, et al:
✓ 3 TCGA cancers & simulated data
✓ cross-validation described in detail
✓ flow diagram
✓ source code & data shared
✓ packages w/ version, cited
/n



✓ 3 TCGA cancers & simulated data
✓ cross-validation described in detail
✓ flow diagram
✓ source code & data shared
✓ packages w/ version, cited
/n
- [the method description & comments follows]
- link: biorxiv.org/content/10.110…
- licence the above figures/tables: CC BY-NC-ND 4.0
- an earlier version of INF was previously presented in 2018: doi.org/10.1186/s13062…
- this is the first tweet in #SundayMultiOmics series
- link: biorxiv.org/content/10.110…
- licence the above figures/tables: CC BY-NC-ND 4.0
- an earlier version of INF was previously presented in 2018: doi.org/10.1186/s13062…
- this is the first tweet in #SundayMultiOmics series
[[Introduction]]: Similarity network fusion (SNF, doi.org/10.1038/nmeth.…) is a popular technique (600+ citations, a lot for multi-omics!) for getting a sort of consensus signal from multiple omics; it requires the same patients (less commonly - observations) in each omic.

