Discover and read the best of Twitter Threads about #scRNAseq
Most recents (24)
New preprint on scGPT: first foundation large language model for single cell biology, pretrained on 10 million cells.
Just as text is made of words, cells are characterized by genes
Some thoughts on how cool this is & why it challenges the status quo of single cell analysis🧵🧵
Just as text is made of words, cells are characterized by genes
Some thoughts on how cool this is & why it challenges the status quo of single cell analysis🧵🧵
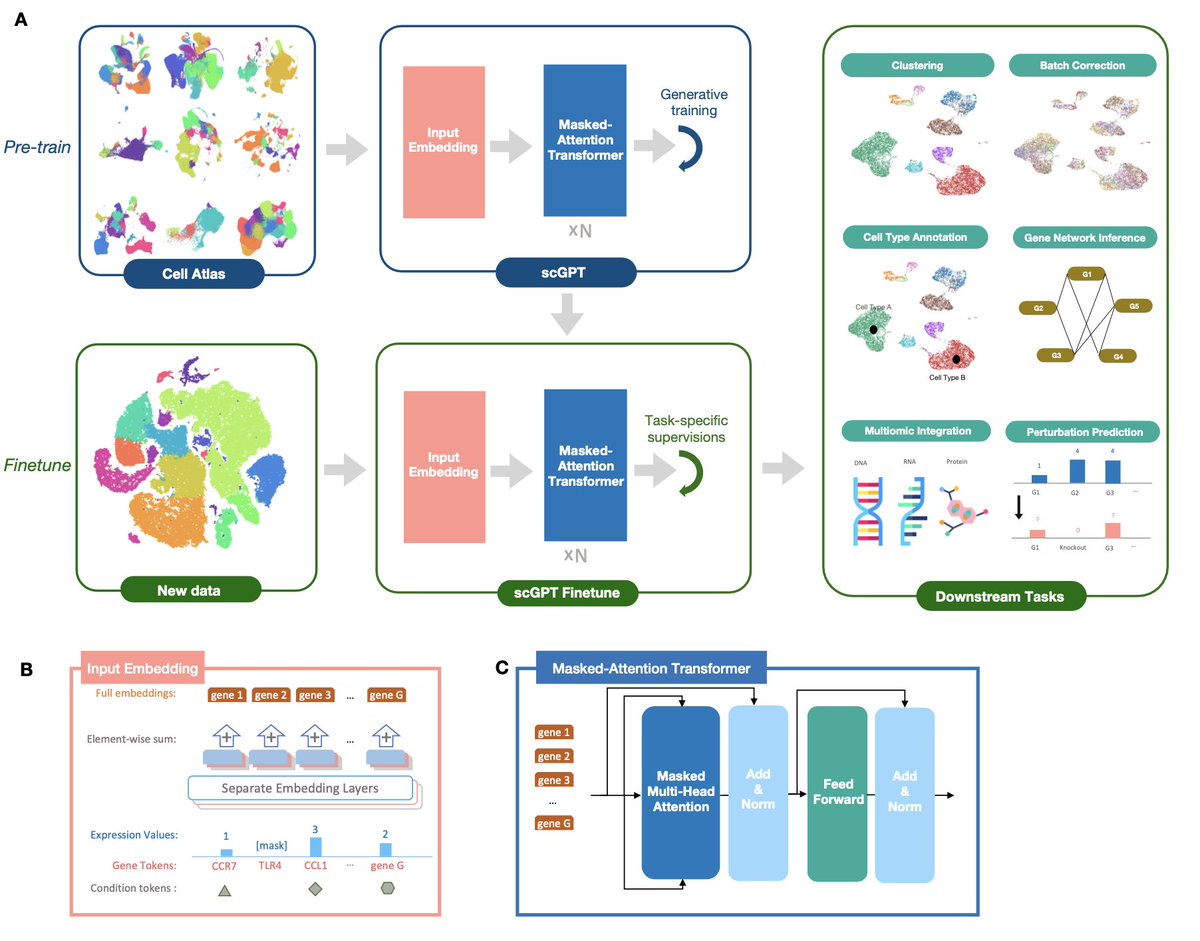
The self-attention transformer (chatGPT) is so successful in natural language processing (NLP)
But maybe single cell biology is not that different from NLP, as genes & cells correspond to words & sentences?
That's the framing of @BoWang87's awesome paper
biorxiv.org/content/10.110…
But maybe single cell biology is not that different from NLP, as genes & cells correspond to words & sentences?
That's the framing of @BoWang87's awesome paper
biorxiv.org/content/10.110…
Another core similarity between NLP & single cell biology is the large and ever-growing size of publicly available #scRNAseq data (e.g human cell atlas) to be used for training.
Can NLP models also understand intrinsic logics of single cell biology & develop "emergent thinking"?
Can NLP models also understand intrinsic logics of single cell biology & develop "emergent thinking"?
Actually, not transforming the data outperforms log(y/s+1). 1/
The "performance" in this analysis boils down to checking consistency of the kNN graph after transformation. That's certainly a property one can optimize for, but it's by no means the only one. In fact, if it was the only property of interest, one could just not transform. 2/
Of course that is trivial and uninteresting. The purpose of normalization is to remove technical noise and stabilize variance. But then one should check how well that is done. And as it turns out, log(y/s+1) actually removes too much "noise". 3/
In a recent preprint with @GorinGennady (biorxiv.org/content/10.110…) we provide a quantitative answer to to this question, namely what information about variance (among cells in a cell type, or more generally many cell types) does a UMAP provide? A short🧵1/
The variability in gene expression across cells can be attributed to biological stochasticity and technical noise. In practice it's hard to break down the variance into these constituent parts. How do we know what is biological vs. technical? 2/
Here's an idea: within a cell type, we can obtain an accurate estimate of gene expression by averaging across cells. Now we can get a lower bound for biological variability by computing the variance across very distinct cell types. 3/
In 2019 "Single-cell multimodal omics" was deemed @naturemethods Method of the Year, and since then many new multimodal methods have been published. But are there tradeoffs w/ multimodal omics?
tl;dr yes! An analysis w/ @sinabooeshaghi & Fan Gao in biorxiv.org/content/10.110… 🧵1/
tl;dr yes! An analysis w/ @sinabooeshaghi & Fan Gao in biorxiv.org/content/10.110… 🧵1/
There are a lot of ways to look at this question and we have much to say (long 🧵ahead!). As a starting point let's begin with our Supplementary Figure 4. This is a comparison of (#snRNAseq+#snATACseq) multimodal technology with unimodal technology. Much to explain here: 2/ 

(a) & (b) are showing the mean-variance relationship for data from an assay for measuring RNA and TAC (transposable accessible chromatin) in the same cells. The data is from ncbi.nlm.nih.gov/geo/query/acc.…
Cells from human HEK293T & mouse NIH3T3 were mixed. You're looking at the RNA. 3/
Cells from human HEK293T & mouse NIH3T3 were mixed. You're looking at the RNA. 3/
Only a matter of time before a paper formalized this exercise:
Automated #scRNAseq cell type annotation with GPT4, evaluated across five datasets, 100s of tissues & cell types, human and mouse.
A🧵below with my thoughts on how such tools will change how #Bioinformatics is done.
Automated #scRNAseq cell type annotation with GPT4, evaluated across five datasets, 100s of tissues & cell types, human and mouse.
A🧵below with my thoughts on how such tools will change how #Bioinformatics is done.

I'll start with a quick summary of the paper, such that we're all on the same page.
(The paper is also a super quick read, literally only 3 pages of text, among which 1 is GPT prompts).
Here's the link to the preprint
biorxiv.org/content/10.110…
(The paper is also a super quick read, literally only 3 pages of text, among which 1 is GPT prompts).
Here's the link to the preprint
biorxiv.org/content/10.110…
The paper looked at 4 already-annotated public datasets: Azimuth, Human Cell Atlas, Human Cell Landscape, Mouse Cell Atlas.
Differentially Expressed Genes characterizing every cluster (DEGe) in these studies were generally available with the publications & were also downloaded.
Differentially Expressed Genes characterizing every cluster (DEGe) in these studies were generally available with the publications & were also downloaded.
I need to raise awareness about an important point in #scRNAseq data analysis, which, in my opinion, is not acknowledged enough:
‼️In practice, most cell type assignment methods will fail on totally novel cell types. Biological/expert curation is necessary!
Here's one example👇
‼️In practice, most cell type assignment methods will fail on totally novel cell types. Biological/expert curation is necessary!
Here's one example👇

Last year, together with @LabPolyak @harvardmed, we published a study in which we did something totally awesome: we experimentally showed how a TGFBR1 inhibitor drug 💊 prevents breast tumor initiation in two different rat models!
Here's a detailed thread on this paper:
Here's a detailed thread on this paper:
Interested in "integrating" multimodal #scRNAseq data? W/ @MariaCarilli, @GorinGennady, @funion10 & Tara Chari we introduce biVI, which combines the scVI variational autoencoder with biophysically motivated bivariate models for RNA distributions. 🧵 1/
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…

One of the clearest cases for "integration" is in combining measurements of nascent and mature mRNAs, which can be obtained with every #scRNAseq experiment. Should "intronic counts" be added to "exonic counts"? Or is it better to pick one or the other? 2/
This important question has been swept under the rug. Perhaps that is because it is inconvenient to have to rethink #scRNAseq with two count matrices as input, instead of one. How does one cluster with two matrices? How does one find marker genes with them? 3/
Bench to bedside series: Lung COPD part 1/3
Respiratory histology (via @drawittoknowit)
Health & COPD Lung @TheLancet
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics

Respiratory histology (via @drawittoknowit)
Health & COPD Lung @TheLancet
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics


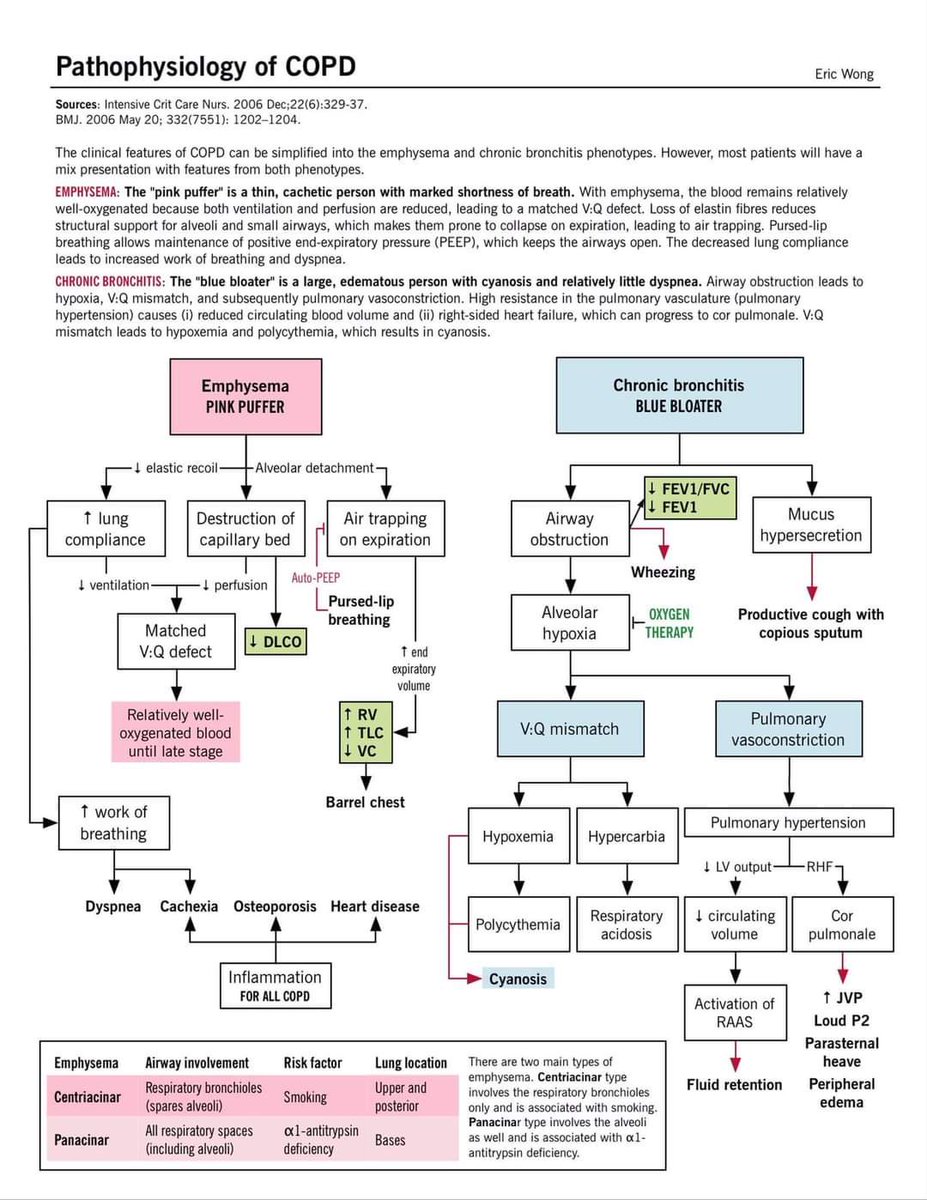
Bench to bedside series: Lung COPD part 2/3
2. #Pathophysiology of #COPD via @EricWong_MD
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics

2. #Pathophysiology of #COPD via @EricWong_MD
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics


Bench to bedside series: Lung COPD part 3/3
#scRNAseq paper: Human distal airways contain a multipotent secretory cell that can regenerate alveoli
1. RASCs (new cell-type) + #stemcell properties in distal airways 2. faulty RASC-to-AT2 transformation in COPD
#Bioinformatics #MedEd
#scRNAseq paper: Human distal airways contain a multipotent secretory cell that can regenerate alveoli
1. RASCs (new cell-type) + #stemcell properties in distal airways 2. faulty RASC-to-AT2 transformation in COPD
#Bioinformatics #MedEd

Healthy Lung vs. Lung with Chronic Obstructive Pulmonary Disease (COPD)
h/t @PatologCritica
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #lung #COPD #INNOMed
h/t @PatologCritica
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #lung #COPD #INNOMed
Bench to bedside series: Lung COPD part 1/3
Respiratory histology (via @drawittoknowit)
Health & COPD Lung @TheLancet
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics

Respiratory histology (via @drawittoknowit)
Health & COPD Lung @TheLancet
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics


Bench to bedside series: Lung COPD part 2/3
2. #Pathophysiology of #COPD via @EricWong_MD
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics
2. #Pathophysiology of #COPD via @EricWong_MD
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics

How Gas Exchange Works
#MedEd #MedTwitter
merckmanuals.com/home/lung-and-…
carolina.com/teacher-resour…

#MedEd #MedTwitter
merckmanuals.com/home/lung-and-…
carolina.com/teacher-resour…


Are you interested in performing splice-aware quantification of your #scrnaseq data, obtaining unspliced, spliced, and ambiguous UMI counts quickly & in <3GB of RAM? If so, check out the new manuscript by @DongzeHe, @CSoneson and me on #bioRxiv bit.ly/3vJr0Ji. 1/🧵
Understanding the origin of sequencing reads — the molecules from which they arise, the "gene" with which those molecules are associated, and the splicing status of those molecules — is a key task in single-cell RNA-seq quantification.
The short, (effectively) single-end nature of the reads used in popular technologies leads to situations in which it can be difficult or impossible to predict if a read was sequenced from an unspliced (nascent) or spliced (mature) RNA; these reads are designated as "ambiguous."
I'm excited to share a paper w co-1st @ar_davino, @TamaraPrietoF + @landau_lab. We introduce tools to study the heritability + plasticity of cell state diversity in somatic evolution, now possible w single-cell tech. It all began with a simple question... biorxiv.org/content/10.110…
#scRNAseq has revealed that across biology, tissues contain lots of cellular diversity, with different groups of cells expressing different sets of genes. But how are these cells ancestrally related? Are cells that look similar also closely related? What would this mean?
To answer these questions, we first need two single-cell measurements, a measure of cell state (eg from scRNAseq) and a measure of ancestry (from lineage tracing). By putting these together, we can see if cells group by both gene expression and ancestry (on a phylogenetic tree).
This flippant comment on #scRNAseq algorithms reflects a common disrespect for computational biologists who are frequently derided for not asking "good biological questions". Moreover, it is peak chutzpah. A short 🧵..
As pointed out by @RArgelaguet, the OP recently coauthored a paper where many #scRNAseq methods, algorithms, and tools were used.. I wonder which of them the OP would have preferred was not developed. @AMartinezArias, please choose from this list:
Bowtie2
nature.com/articles/nmeth…
nature.com/articles/nmeth…
Our immune system is essential in keeping us healthy.
But the immune system also changes profoundly as we age.
Why is that? Could we prevent it?
Let's see how #singlecell biology can help us better understand #immune #aging
🧵👇
But the immune system also changes profoundly as we age.
Why is that? Could we prevent it?
Let's see how #singlecell biology can help us better understand #immune #aging
🧵👇
First, some background.
Everybody knows that the immune system is hugely complex.
#singlecell sequencing has (arguably) done more for the immune system than for other health applications.
Via #scRNAseq, we discovered & characterized crazily detailed immune cell phenotypes.
Everybody knows that the immune system is hugely complex.
#singlecell sequencing has (arguably) done more for the immune system than for other health applications.
Via #scRNAseq, we discovered & characterized crazily detailed immune cell phenotypes.

Such detailed phenotypes have been found in both healthy and diseased tissues.
I wrote several threads about this topic and find it to be one of the most foundational & fascinating progresses that have happened in biomedicine in the past 10 years.
I wrote several threads about this topic and find it to be one of the most foundational & fascinating progresses that have happened in biomedicine in the past 10 years.
#singlecell analysis is revolutionizing medicine and changing the way we look at disease.
New perspective article just out🚨@NatureMedicine reflecting on @humancellatlas: informative for both #singlecell lovers❤️& skeptiks🤔
Let's map out where the field stands & what is next🧵
New perspective article just out🚨@NatureMedicine reflecting on @humancellatlas: informative for both #singlecell lovers❤️& skeptiks🤔
Let's map out where the field stands & what is next🧵

First, some context.
The genomics single cell field has started out 1-2 decades ago with a huge promise:
"Find the missing link between genes, diseases and therapies. This will bring completely novel therapeutics to the market & cure disease."
The genomics single cell field has started out 1-2 decades ago with a huge promise:
"Find the missing link between genes, diseases and therapies. This will bring completely novel therapeutics to the market & cure disease."
The underlying logic is straigtforward:
1. the cell is the main unit of living organisms
⬇️
2. cells break down in disease
⬇️
3. understanding cells helps understand how & why they break
⬇️
4. this helps with engineering new therapeutics
⬇️
5. new therapeutics will cure disease
1. the cell is the main unit of living organisms
⬇️
2. cells break down in disease
⬇️
3. understanding cells helps understand how & why they break
⬇️
4. this helps with engineering new therapeutics
⬇️
5. new therapeutics will cure disease
Can we outsmart #cancer and stop it before it even starts?
Our brand new paper🔥@NatureComms reveals a novel stem-like cell population directly related to #breast tumor initiation.
Let's dig in🧵🧵
Our brand new paper🔥@NatureComms reveals a novel stem-like cell population directly related to #breast tumor initiation.
Let's dig in🧵🧵

First, quick background.
Sadly, everybody reading this knows breast cancer.
It is the most commonly diagnosed cancer in women, with a staggering 1 in every 8 women in the world receiving this diagnosis throughout their lifetimes.
Sadly, everybody reading this knows breast cancer.
It is the most commonly diagnosed cancer in women, with a staggering 1 in every 8 women in the world receiving this diagnosis throughout their lifetimes.
In a new preprint w/@kreldjarn, @DelaneyKSull, @GuillaumOleSan & @pmelsted we address a shortcoming in current approaches to quantifying single-nucleus RNA-seq: biorxiv.org/content/10.110…
tl;dr care has to be taken in quantifying nascent vs. mature transcripts. #snRNAseq #scRNAseq🧵
tl;dr care has to be taken in quantifying nascent vs. mature transcripts. #snRNAseq #scRNAseq🧵
First, note that single-cell RNA-seq data provides some quantification not only of processed (mature) messenger RNAs, but also of nascent molecules. That observation by @GioeleLaManno, @slinnarsson and colleagues underlies RNA velocity. But what about single-nucleus RNA-seq? 2/
New🔥 #DataScience #Bioinformatics resource: 850,000‼️ #scRNAseq cells from 226 samples across 10 cancer types draw a map of the tumor microenvironment, in particular fibroblasts.
Let’s see👇what are the main contributions of this work & what this means for #cancer #Genomics🧵
Let’s see👇what are the main contributions of this work & what this means for #cancer #Genomics🧵
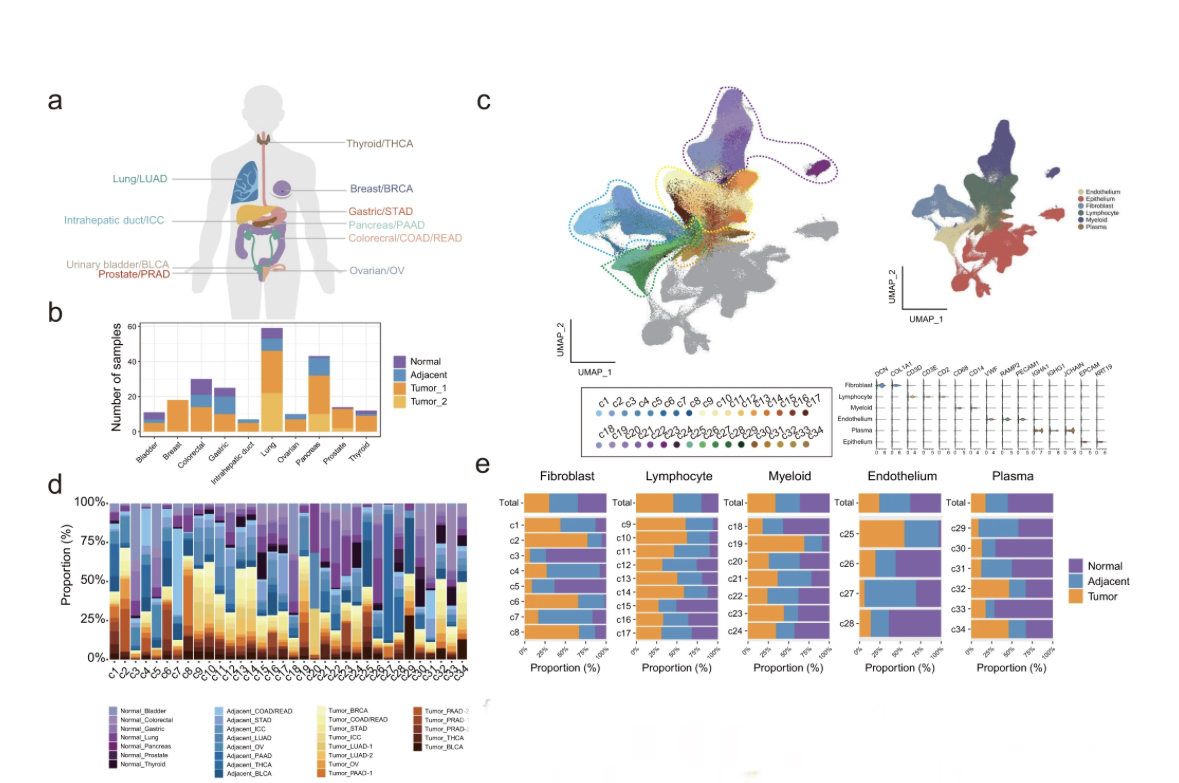
But first, some background.
Cancers are (unfortunately) complex ecosystems,consisting of various types of cells.
Malignant cells represent only a fraction of the tumor. The rest is made of the tumor microenvironment/TME (fibroblasts + immune cells), with complicated dual roles.
Cancers are (unfortunately) complex ecosystems,consisting of various types of cells.
Malignant cells represent only a fraction of the tumor. The rest is made of the tumor microenvironment/TME (fibroblasts + immune cells), with complicated dual roles.

Understanding the essence of this duality is key in understanding why most cancer therapies fail.
TME cells are plastic & can easily change states.
The same TME cells can either promote or suppress tumor development, depending on very subtle factors totally not well understood.
TME cells are plastic & can easily change states.
The same TME cells can either promote or suppress tumor development, depending on very subtle factors totally not well understood.
Amazing week for #DeepLearning in #spatial #singlecell biology, with 2🔥new Graph Neural Networks methods!
1.STELLAR🇺🇸 @jure: a cell type annotation & discovery atlas-type framework
2.NCEM🇪🇺 @fabian_theis: an approach to infer cellular communication patterns
Deep dive below🧵
1.STELLAR🇺🇸 @jure: a cell type annotation & discovery atlas-type framework
2.NCEM🇪🇺 @fabian_theis: an approach to infer cellular communication patterns
Deep dive below🧵

But first, some background.
Spatial molecular biology has actually been around since the 70s. @lpachter's wonderful book-like article "Museum of Spatial Transcriptomics" comprehensively discusses history, tech & methodology advances in the past 50 years.
nature.com/articles/s4159…
Spatial molecular biology has actually been around since the 70s. @lpachter's wonderful book-like article "Museum of Spatial Transcriptomics" comprehensively discusses history, tech & methodology advances in the past 50 years.
nature.com/articles/s4159…
Nevertheless, recent advances in single cell molecular technologies (brought by e.g. @10xGenomics & @AkoyaBio) have facilitated the high-throughout profiling of (groups of) single cells in their tissue context across embryogenesis, normal tissue development & disease progression.
Today's amazing science dives deep into the 2 strongest #cancer modulators: evolution & immune defense.
First-ever detailed temporal evolutionary trajectories for 600,000 B cell lymphoma immune cells #scRNAseq & #scTCRSeq of 32 patients during immunotherapy with 2 CAR-T drugs 🧵
First-ever detailed temporal evolutionary trajectories for 600,000 B cell lymphoma immune cells #scRNAseq & #scTCRSeq of 32 patients during immunotherapy with 2 CAR-T drugs 🧵

First, what is chimeric antigen receptor (CAR) T cell therapy?
It is an immunotherapy in which the patient's own immune cells are genetically engineered ex-vivo to recognize, attack & kill tumor cells. Then they are infused back into the patient, ready to fight the enemy!🤺 2/13
It is an immunotherapy in which the patient's own immune cells are genetically engineered ex-vivo to recognize, attack & kill tumor cells. Then they are infused back into the patient, ready to fight the enemy!🤺 2/13
Immunotherapies have revolutionized cancer treatment & are among the most promising future approaches.
However: response rates, even if varying across cancers, remain limited, with e.g. 50% response in lymphomas.
Why such therapies fail for the other half remains a mystery 3/13
However: response rates, even if varying across cancers, remain limited, with e.g. 50% response in lymphomas.
Why such therapies fail for the other half remains a mystery 3/13
Valuable #immunology cell atlas: #scRNAseq + paired B & T cell receptor seq for 330,000 tissue-resident immune cells across 16 human tissues.
CellTypist: new & robust immune cells annotation algorithm, finding 101 immune cell types in 1,000,000 cells‼️
Why this is important👇🧵
CellTypist: new & robust immune cells annotation algorithm, finding 101 immune cell types in 1,000,000 cells‼️
Why this is important👇🧵

How wounds heal
#woundhealing #injury #homeostasis #angiogenesis #epidermal #skin #MedTwitter #meded #foamed #InnoMed
#woundhealing #injury #homeostasis #angiogenesis #epidermal #skin #MedTwitter #meded #foamed #InnoMed
Timelapse Of A Wound Healing
#epidermal #medtwitter #meded #foamed #InnoMed #homeostasis #injuy #woundhealing #sciencetwitter
#epidermal #medtwitter #meded #foamed #InnoMed #homeostasis #injuy #woundhealing #sciencetwitter
How does #aging affect #WoundHealing?
#scRNAseq identifies major changes in cell compositions, kinetics, and molecular profiles during #woundhealing in aged #skin @CellReports
bit.ly/3PZMies
#epidermal #MedTwitter #meded #foamed #InnoMed #bioinformatics #homeostasis
#scRNAseq identifies major changes in cell compositions, kinetics, and molecular profiles during #woundhealing in aged #skin @CellReports
bit.ly/3PZMies
#epidermal #MedTwitter #meded #foamed #InnoMed #bioinformatics #homeostasis

Bench to bed series: Lung COPD part 1/3
1. #Respiratory #histology (via @drawittoknowit)
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics
1. #Respiratory #histology (via @drawittoknowit)
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics

Bench to bed series: Lung COPD part 2/3
2. #Pathophysiology of #COPD via @EricWong_MD
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics
2. #Pathophysiology of #COPD via @EricWong_MD
#4KMedEd #meded #foamed #medtwitter #MedEd #MedTwitter #Pulmtwitter #scRNAseq #Bioinformatics
Bench to bed series: Lung COPD part 3/3
#scRNAseq paper: Human distal airways contain a multipotent secretory cell that can regenerate alveoli
1. RASCs (new cell-type) + #stemcell properties in distal airways 2. faulty RASC-to-AT2 transformation in COPD
#Bioinformatics #meded
#scRNAseq paper: Human distal airways contain a multipotent secretory cell that can regenerate alveoli
1. RASCs (new cell-type) + #stemcell properties in distal airways 2. faulty RASC-to-AT2 transformation in COPD
#Bioinformatics #meded

For centuries we've had anatomical maps of how the body's organs are connected, but what would a diagram of the immune system’s connections look like? In @Nature we report our initiative to map the "interactome" that links human immune cells together (1/14)nature.com/articles/s4158… 
The life of an immune cell is constantly on the move. As white blood cells (leukocytes) circulate throughout the body, they must dynamically form connections with each other in order to communicate messages like “threat detected!” or “stop attacking, this is healthy tissue”(2/14)
Leukocytes can physically interact through #receptor proteins on their surfaces that have evolved to recognize and bind each other. These receptor interactions have enormous medical significance, to cite the #immuntherapy revolution in cancer treatment as just one example (3/14) 





